Differences between Python and Julia
The IAI Python interface matches the Julia API very closely, so you can refer to the Julia documentation for information on most tasks. On this page we note the main differences between the Python and Julia interfaces.
Conversion of Julia data types to Python
In order to figure out the types to pass to an IAI function from the Python interface, you can refer to the equivalent function in the Julia API and translate the types to their Python equivalent. Most literal data types convert in a straighforward manner, for example:
InttointFloat64tofloatStringtostrDicttodict
The following Julia types can be passed as follows:
nothingcan be passed usingNone- a
Symbolcan be passed as astr - a
Vectorcan be passed as alist, 1-Dnumpy.arrayor apandas.Series - a
Matrixcan be passed as a 2-Dnumpy.array - a
DataFramecan be passed as apandas.DataFrame
Specifying Feature Set in Python
We list the Python input types for specifying set of features in a dataframe as learner parameters. Refer to IAI.FeatureSet for the Julia equivalence. Note that if you are using integers to specify the indices of columns, the input is expected to use one-based indexing as in Julia.
| Input Type | Description | Examples |
|---|---|---|
All | Use all columns | {'All'}, {'All' : []} |
Integer or a vector of Integers | Specify indices of columns to use | 1, [1, 3, 4] |
String or a vector of Strings | Specify names of columns to use | 'x1', ['x1', 'x3'] |
Not | Specify columns not to use | {'Not' : 1}, {'Not' : ['x2', 'x4']} |
Between | Specify range of columns to use | {'Between' : ['x1', 'x4']} |
Object-oriented interface for learners
In the IAI Python interface, the API methods relating to learners are methods of the learner objects rather than functions that operate on learners as in the Julia interface. For instance the IAI.fit! method in Julia:
IAI.fit!(lnr, X, y)would be called from the Python interface as
lnr.fit(X, y)Interactive Visualizations
The write_html and show_in_browser functions work the same in Python as in Julia for saving visualizations to file or displaying in an external browser, respectively. Additionally, visualizations will be automatically shown in Jupyter notebooks as they are for Julia.
Below is an example that shows the equivalent Python code for the advanced visualization examples in Julia. In these examples we work with the following tree learner:
We can rename the features with a dict that maps from the original names to more descriptive names:
vis_renamed_features = lnr.TreePlot(feature_renames={
"Disp": "Displacement",
"HP": "Horsepower",
"WT": "Weight",
})
We can also have a finer-grained control of what is displayed for each node, such as adding summary statistics. We create a list of dicts with the parameters controlling what you want to show in each node and pass this as extra_content:
import numpy as np
node_inds = lnr.apply_nodes(X)
def get_text(inds):
content = ('<b>Mean horsepower in node:</b> ' +
str(np.round(np.mean(X.HP.iloc[inds]), decimals=2)))
return ({'node_details_extra' : content})
extras = [get_text(inds) for inds in node_inds]
vis_extra_text = lnr.TreePlot(extra_content=extras)
Finally, we can combine multiple learners in a single visualization as described in the Julia documentation. In Python, a question is a tuple of the form (question, responses), where question is the string prompt for the question and responses is a list of possible responses:
questions = ("Use learner with", [
("renamed features", vis_renamed_features),
("extra text output", vis_extra_text)
])
iai.MultiTreePlot(questions)
Tree Stability
Below are examples showing the equivalent Python code for the tree stability examples in Julia. In these examples we work with the following tree learner:
Stability Analysis
We conduct the stability analysis using StabilityAnalysis:
stability = iai.StabilityAnalysis(lnr, train_X, train_y, criterion='gini')
We can plot a summary of the analysis using plot:
stability.plot()
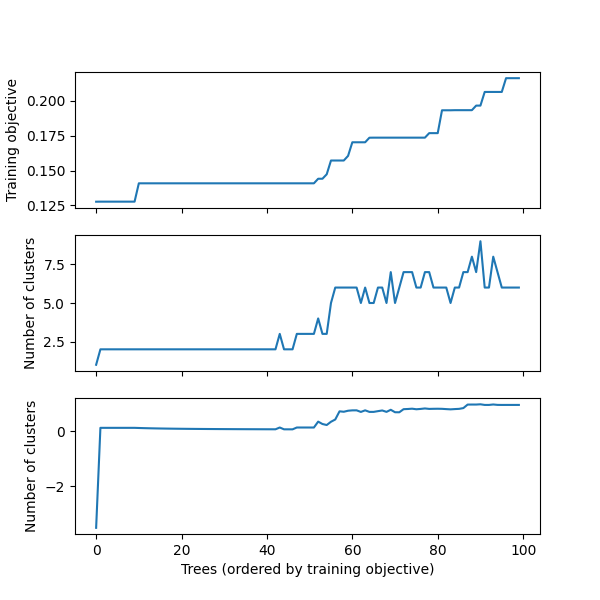
We can use get_stability_results to extract the trees in order of training objective along with the importance of each feature in the tree:
stability.get_stability_results()
train_error tree_index variance skewness curtosis entropy
0 0.127639 38 0.601713 0.395491 0.002795 0.0
1 0.127677 21 0.615924 0.237532 0.146544 0.0
2 0.127677 22 0.615924 0.237532 0.146544 0.0
3 0.127677 29 0.615924 0.237532 0.146544 0.0
4 0.127677 39 0.615924 0.237532 0.146544 0.0
5 0.127677 55 0.615924 0.237532 0.146544 0.0
6 0.127677 59 0.615924 0.237532 0.146544 0.0
.. ... ... ... ... ... ...
93 0.206366 36 0.783420 0.185882 0.030698 0.0
94 0.206366 57 0.783420 0.185882 0.030698 0.0
95 0.206366 76 0.783420 0.185882 0.030698 0.0
96 0.216240 87 0.746042 0.195427 0.058531 0.0
97 0.216240 25 0.764514 0.196132 0.039354 0.0
98 0.216240 30 0.764514 0.196132 0.039354 0.0
99 0.216240 100 0.764514 0.196132 0.039354 0.0
[100 rows x 6 columns]We can use get_cluster_details to summarize the clustering of the first 10 trees:
stability.get_cluster_details(10)
train_error_mean variance skewness curtosis entropy
0 0.127639 0.601713 0.395491 0.002795 0.0
1 0.127677 0.615924 0.237532 0.146544 0.0We can use get_cluster_distances to get the relative distances between each pair of clusters:
stability.get_cluster_distances(10)
array([[0. , 0.21404831],
[0.21404831, 0. ]])We can use get_cluster_assignments to see which trees comprise each cluster:
stability.get_cluster_assignments(10)
[array([38], dtype=int64), array([21, 22, 29, 39, 55, 59, 70, 96, 97], dtype=int64)]We see that Tree 38 is in its own cluster, and the other trees are all grouped together. Given this, we might want to inspect how Tree 38 differs to the others. We can use get_tree to construct a new learner that uses the tree at a specified index:
lnr.get_tree(38)
We can also generate an interactive visualization that allows us to explore all of the information in the stability analysis using write_html or show_in_browser:
stability.write_html("stability_analysis.html")
The resulting interactive visualization can be viewed here. As with all visualizations, this visualization is also automatically shown inside a Jupyter notebook.
Similarity Comparison
We can also measure the stability of the training process to changes in the training data. We will use the following learner, trained on a variation of the training data:
We first use variable_importance_similarity to compute the difference between the original tree and each tree in the new learner, and feed this into SimilarityComparison:
deviations = lnr.variable_importance_similarity(new_lnr, test_X, test_y,
criterion='gini')
similarity = iai.SimilarityComparison(lnr, new_lnr, deviations)
We can plot a summary of the analysis using plot:
similarity.plot()
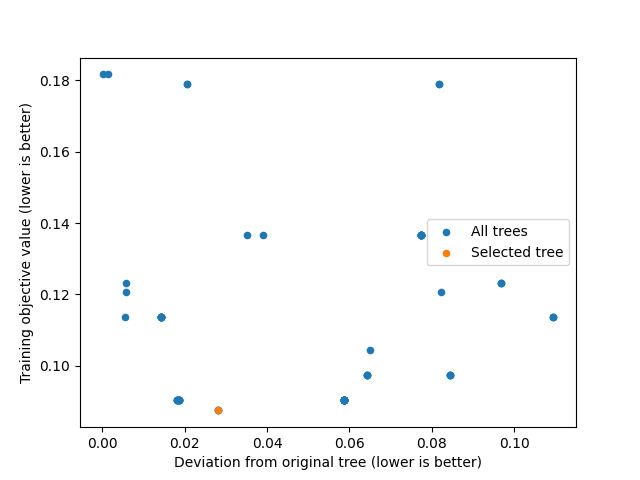
We can get the training objective values with get_train_errors:
train_errors = similarity.get_train_errors()
array([0.12311935, 0.09033247, 0.09033247, ..., 0.11373525, 0.09033247,
0.17885706])As we did for stability analysis, we can also generate an interactive visualization that allows us to explore all of the information in the similarity comparison using write_html or show_in_browser:
similarity.write_html("similarity_comparison.html")
The resulting interactive visualization can be viewed here. As with all visualizations, this visualization is also automatically shown inside a Jupyter notebook.